|
chr11_-_55223449
|
102.320
|
|
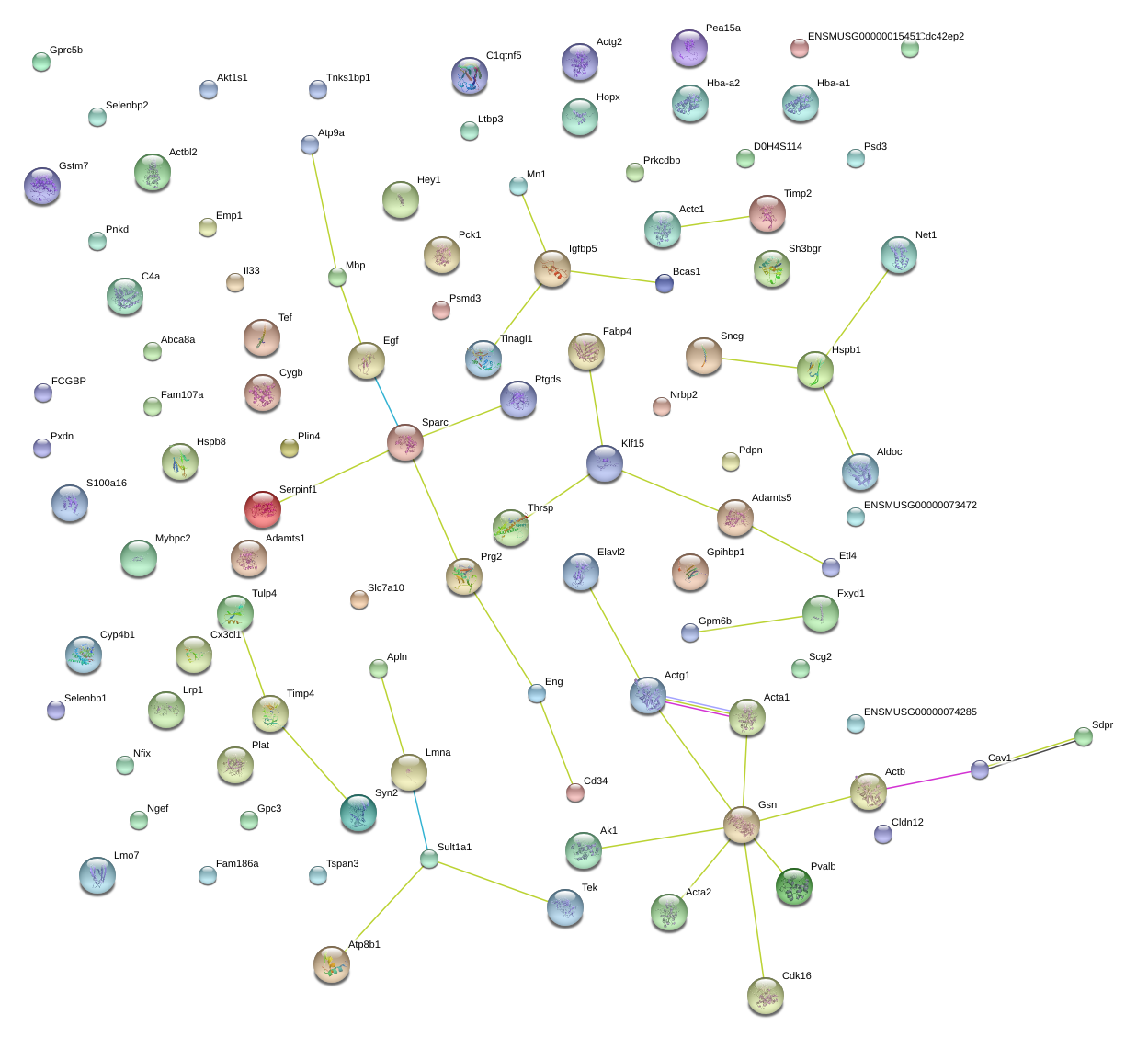
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr18_-_33623069
|
87.230
|
NM_001109990
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr14_+_102237708
|
75.775
|
|
Lmo7
|
LIM domain only 7
|
|
chr18_-_33623668
|
75.088
|
NM_001109988
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr6_+_17256383
|
66.222
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr18_-_33623287
|
66.192
|
NM_053078
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr18_-_33623400
|
65.714
|
NM_001109989
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr6_+_17256415
|
65.080
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr6_+_17256369
|
64.906
|
NM_007616
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr19_+_29999596
|
58.320
|
NM_133775
|
Il33
|
interleukin 33
|
|
chr1_+_51345952
|
55.084
|
NM_138741
|
Sdpr
|
serum deprivation response
|
|
chr3_+_90341208
|
51.572
|
|
S100a16
|
S100 calcium binding protein A16
|
|
chr8_-_87323896
|
50.963
|
|
Nfix
|
nuclear factor I/X
|
|
chr2_-_25325196
|
50.935
|
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr1_+_51346086
|
50.927
|
|
Sdpr
|
serum deprivation response
|
|
chr5_-_116872852
|
50.512
|
NM_030704
|
Hspb8
|
heat shock protein 8
|
|
chr11_-_116515602
|
49.383
|
NM_030206
|
Cygb
|
cytoglobin
|
|
chr1_-_89470442
|
48.599
|
NM_001111314
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr4_-_115320309
|
48.141
|
NM_007823
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1
|
|
chr1_+_51346108
|
47.129
|
|
Sdpr
|
serum deprivation response
|
|
chr7_-_133819870
|
45.381
|
NM_133670
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1
|
|
chr15_-_78034648
|
44.803
|
|
Pvalb
|
parvalbumin
|
|
chr2_-_25325248
|
44.702
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr5_+_136363769
|
44.579
|
NM_013560
|
Hspb1
|
heat shock protein 1
|
|
chr18_+_82723920
|
44.575
|
|
Mbp
|
myelin basic protein
|
|
chr16_+_96422243
|
44.439
|
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr18_+_82723854
|
44.159
|
NM_001025251
NM_001025255
NM_001025256
NM_001025258
NM_001025259
NM_001025254
|
Mbp
|
myelin basic protein
|
|
chr18_+_82723907
|
43.535
|
|
Mbp
|
myelin basic protein
|
|
chr11_-_116515361
|
43.187
|
|
Cygb
|
cytoglobin
|
|
chr15_-_78034565
|
42.813
|
NM_013645
|
Pvalb
|
parvalbumin
|
|
chr10_-_127058163
|
42.667
|
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chr11_-_118216819
|
42.036
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr11_-_118216625
|
40.118
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr3_-_10208575
|
39.103
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr11_-_118216689
|
38.724
|
NM_011594
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr2_+_32483909
|
38.310
|
NM_001198792
|
Ak1
|
adenylate kinase 1
|
|
chrX_+_162676903
|
38.208
|
|
Gpm6b
|
glycoprotein m6b
|
|
chr7_+_35971403
|
37.546
|
NM_017394
|
Slc7a10
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 10
|
|
chr11_-_118216591
|
36.704
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr5_+_136363810
|
36.277
|
|
Hspb1
|
heat shock protein 1
|
|
chr2_-_168559743
|
36.105
|
|
Atp9a
|
ATPase, class II, type 9A
|
|
chr5_+_136363914
|
35.784
|
|
Hspb1
|
heat shock protein 1
|
|
chr1_+_74379182
|
34.647
|
NM_019999
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia
|
|
chr3_+_94736993
|
34.501
|
NM_009150
|
Selenbp1
|
selenium binding protein 1
|
|
chr9_-_56008770
|
34.374
|
|
Tspan3
|
tetraspanin 3
|
|
chr8_+_23868194
|
34.252
|
NM_008872
|
Plat
|
plasminogen activator, tissue
|
|
chr16_-_85803359
|
33.897
|
NM_009621
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1
|
|
chr1_-_72921438
|
33.539
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr9_-_56008819
|
33.158
|
|
Tspan3
|
tetraspanin 3
|
|
chr5_-_5514787
|
32.509
|
NM_001193660
NM_001193659
NM_001193661
NM_022890
|
Cldn12
|
claudin 12
|
|
chr3_-_8666930
|
32.079
|
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr3_+_94736974
|
32.062
|
|
Selenbp1
|
selenium binding protein 1
|
|
chr14_-_9142289
|
31.962
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr9_-_56008595
|
31.068
|
|
Tspan3
|
tetraspanin 3
|
|
chr9_-_56008828
|
29.945
|
NM_019793
|
Tspan3
|
tetraspanin 3
|
|
chr11_-_109957250
|
29.701
|
NM_153145
|
Abca8a
|
ATP-binding cassette, sub-family A (ABC1), member 8a
|
|
chr17_-_56249206
|
29.544
|
NM_020568
|
Plin4
|
perilipin 4
|
|
chr5_-_5514729
|
29.310
|
|
Cldn12
|
claudin 12
|
|
chr7_-_31839825
|
29.206
|
NM_052991
NM_194321
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr11_+_78137699
|
29.165
|
NM_009657
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr2_+_20211539
|
28.884
|
NM_001081006
|
Etl4
|
enhancer trap locus 4
|
|
chrX_+_20265093
|
28.821
|
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr1_-_79436564
|
28.694
|
NM_009129
|
Scg2
|
secretogranin II
|
|
chr16_-_85803343
|
28.092
|
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1
|
|
chr15_-_75921924
|
27.869
|
|
Nrbp2
|
nuclear receptor binding protein 2
|
|
chr7_-_126138689
|
27.564
|
NM_001195774
NM_022420
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr5_-_77544147
|
27.473
|
NM_175606
|
Hopx
|
HOP homeobox
|
|
chr1_+_196765014
|
27.163
|
NM_001111059
NM_133654
|
Cd34
|
CD34 antigen
|
|
chr2_-_113878535
|
26.773
|
NM_009608
|
Actc1
|
actin, alpha, cardiac muscle 1
|
|
chr6_-_115201866
|
25.956
|
NM_080639
|
Timp4
|
tissue inhibitor of metalloproteinase 4
|
|
chr12_+_30687597
|
25.598
|
|
Pxdn
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_72921098
|
25.440
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr7_-_104566013
|
25.118
|
NM_009381
|
Thrsp
|
thyroid hormone responsive SPOT14 homolog (Rattus)
|
|
chr6_+_90424727
|
24.951
|
|
Klf15
|
Kruppel-like factor 15
|
|
chr17_+_6106829
|
24.811
|
NM_001103181
|
Tulp4
|
tubby like protein 4
|
|
chr2_+_35122282
|
24.439
|
|
Gsn
|
gelsolin
|
|
chr1_+_196765079
|
24.405
|
|
Cd34
|
CD34 antigen
|
|
chr3_-_8666985
|
24.267
|
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr15_+_81633192
|
23.646
|
|
Tef
|
thyrotroph embryonic factor
|
|
chr10_-_126978704
|
23.279
|
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chrX_+_20265584
|
22.996
|
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr6_-_115201846
|
22.833
|
|
Timp4
|
tissue inhibitor of metalloproteinase 4
|
|
chr19_+_5741862
|
22.759
|
|
Ltbp3
|
latent transforming growth factor beta binding protein 3
|
|
chr17_-_34960292
|
22.728
|
NM_011413
|
C4a
|
complement component 4A (Rodgers blood group)
|
|
chr15_+_75427044
|
22.077
|
NM_026730
|
Gpihbp1
|
GPI-anchored HDL-binding protein 1
|
|
chr19_+_29999624
|
22.037
|
|
Il33
|
interleukin 33
|
|
chr8_-_126418628
|
22.007
|
NM_009606
|
Acta1
|
actin, alpha 1, skeletal muscle
|
|
chrX_-_49967112
|
21.992
|
NM_016697
|
Gpc3
|
glypican 3
|
|
chr2_+_32502107
|
21.886
|
NM_001146348
NM_001146350
NM_007932
|
Eng
|
endoglin
|
|
chr7_-_51780003
|
21.816
|
NM_146189
|
Mybpc2
|
myosin binding protein C, fast-type
|
|
chr3_-_129458190
|
21.699
|
NM_010113
|
Egf
|
epidermal growth factor
|
|
chr9_+_43915391
|
21.684
|
NM_001190319
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5
|
|
chr4_-_142889372
|
21.525
|
NM_010329
|
Pdpn
|
podoplanin
|
|
chr4_-_129852325
|
21.440
|
NM_023476
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr16_+_96422285
|
21.267
|
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr8_+_97296066
|
21.183
|
NM_009142
|
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_-_77543903
|
21.138
|
|
Hopx
|
HOP homeobox
|
|
chr7_-_112630662
|
20.867
|
NM_028444
|
Prkcdbp
|
protein kinase C, delta binding protein
|
|
chr14_-_35187767
|
20.718
|
NM_011430
|
Sncg
|
synuclein, gamma
|
|
chr9_+_43915327
|
20.690
|
NM_001040631
NM_001040632
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5
|
|
chr18_-_64820573
|
20.532
|
NM_001001488
|
Atp8b1
|
ATPase, class I, type 8B, member 1
|
|
chr2_+_172978566
|
20.384
|
NM_011044
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic
|
|
chr3_-_88297220
|
20.284
|
NM_019390
|
Lmna
|
lamin A
|
|
chrX_-_49967090
|
20.175
|
|
Gpc3
|
glypican 3
|
|
chr6_+_135313010
|
20.152
|
|
Emp1
|
epithelial membrane protein 1
|
|
chr13_-_3917356
|
19.868
|
NM_019671
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr7_-_126138646
|
19.740
|
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr3_+_94497485
|
19.688
|
NM_019414
|
Selenbp2
|
selenium binding protein 2
|
|
chr1_+_196765124
|
19.506
|
|
Cd34
|
CD34 antigen
|
|
chr1_-_174136874
|
19.371
|
NM_011063
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr4_+_94405963
|
19.333
|
NM_013690
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr7_+_52105323
|
19.244
|
|
Akt1s1
|
AKT1 substrate 1 (proline-rich)
|
|
chr7_+_52104591
|
18.992
|
NM_026270
|
Akt1s1
|
AKT1 substrate 1 (proline-rich)
|
|
chr7_+_28914482
|
18.975
|
NM_001164655
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene
|
|
chr3_-_8667013
|
18.766
|
NM_010423
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr2_+_84890611
|
18.739
|
NM_001081260
|
Tnks1bp1
|
tankyrase 1 binding protein 1
|
|
chr8_-_70342105
|
18.631
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr5_+_111847185
|
18.560
|
NM_001081235
|
Mn1
|
meningioma 1
|
|
chr11_+_32196488
|
18.536
|
NM_001083955
NM_008218
|
Hba-a2
Hba-a1
|
hemoglobin alpha, adult chain 2
hemoglobin alpha, adult chain 1
|
|
chr2_-_170232008
|
18.513
|
|
Bcas1
|
breast carcinoma amplified sequence 1
|
|
chr11_-_75236085
|
18.466
|
NM_011340
|
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1
|
|
chr19_-_5924796
|
18.429
|
|
Cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chrX_-_45388010
|
18.250
|
NM_013912
|
Apln
|
apelin
|
|
chr16_+_96422076
|
18.035
|
NM_015825
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr4_-_91038729
|
17.972
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr3_-_107734481
|
17.856
|
NM_026672
|
Gstm7
|
glutathione S-transferase, mu 7
|
|
chr2_+_32502186
|
17.811
|
|
Eng
|
endoglin
|
|
chr7_-_26180629
|
17.796
|
|
Lipe
|
lipase, hormone sensitive
|
|
chr12_+_96930429
|
17.779
|
NM_201518
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr13_+_38436987
|
17.639
|
|
Bmp6
|
bone morphogenetic protein 6
|
|
chr2_-_115890747
|
17.231
|
NM_001136072
NM_001159567
NM_001159568
NM_010825
|
Meis2
|
Meis homeobox 2
|
|
chr3_-_83967952
|
17.226
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr10_+_115738429
|
17.207
|
NM_029928
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr13_-_69138164
|
17.101
|
|
Adcy2
|
adenylate cyclase 2
|
|
chr6_+_135312948
|
17.072
|
NM_010128
|
Emp1
|
epithelial membrane protein 1
|
|
chr15_+_81633102
|
16.779
|
NM_153484
|
Tef
|
thyrotroph embryonic factor
|
|
chr4_-_150482562
|
16.543
|
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr17_-_56261311
|
16.425
|
NM_029796
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr1_-_174136809
|
16.057
|
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr5_+_21049296
|
16.043
|
NM_028977
|
Lrrc17
|
leucine rich repeat containing 17
|
|
chr6_+_17231334
|
15.988
|
NM_016900
|
Cav2
|
caveolin 2
|
|
chr14_+_121434765
|
15.877
|
NM_134082
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr7_+_147960463
|
15.794
|
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr6_-_55124983
|
15.742
|
NM_009349
|
Inmt
|
indolethylamine N-methyltransferase
|
|
chr2_-_25978328
|
15.281
|
NM_026495
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing
|
|
chr1_+_132723258
|
15.229
|
NM_011082
|
Pigr
|
polymeric immunoglobulin receptor
|
|
chr11_+_78136681
|
15.206
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr7_+_138175581
|
15.089
|
NM_007769
|
Dmbt1
|
deleted in malignant brain tumors 1
|
|
chr16_-_84835213
|
15.010
|
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F
|
|
chr4_+_62360705
|
14.927
|
|
Rgs3
|
regulator of G-protein signaling 3
|
|
chr13_+_49639799
|
14.870
|
NM_001172481
NM_025711
|
Aspn
|
asporin
|
|
chr19_+_32694494
|
14.787
|
NM_011864
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr11_-_101026399
|
14.648
|
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr7_-_108025317
|
14.647
|
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr4_-_57929087
|
14.594
|
NM_178727
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene
|
|
chr16_-_18426802
|
14.567
|
NM_001111062
NM_007744
|
Comt
|
catechol-O-methyltransferase
|
|
chr7_-_20355818
|
14.564
|
|
Bcam
|
basal cell adhesion molecule
|
|
chr7_-_28118628
|
14.543
|
NM_001113549
|
Ltbp4
|
latent transforming growth factor beta binding protein 4
|
|
chr11_+_28753190
|
14.485
|
NM_146015
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1
|
|
chr2_+_106533615
|
14.331
|
NM_001143683
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr15_+_54577256
|
14.196
|
NM_010930
|
Nov
|
nephroblastoma overexpressed gene
|
|
chr2_+_106533281
|
14.193
|
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr6_-_115201790
|
13.895
|
|
Timp4
|
tissue inhibitor of metalloproteinase 4
|
|
chr11_-_75236012
|
13.817
|
|
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1
|
|
chr10_+_57504720
|
13.742
|
NM_021272
|
Fabp7
|
fatty acid binding protein 7, brain
|
|
chr7_+_138079669
|
13.703
|
NM_019564
|
Htra1
|
HtrA serine peptidase 1
|
|
chr8_-_49760037
|
13.687
|
NM_001145937
NM_011857
|
Odz3
|
odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr8_+_91115856
|
13.595
|
|
Nkd1
|
naked cuticle 1 homolog (Drosophila)
|
|
chr8_-_37796104
|
13.359
|
NM_001194941
|
Dlc1
|
deleted in liver cancer 1
|
|
chr1_+_132723277
|
13.281
|
|
Pigr
|
polymeric immunoglobulin receptor
|
|
chr1_+_43787692
|
13.141
|
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr2_+_25145414
|
13.127
|
NM_146117
|
Lrrc26
|
leucine rich repeat containing 26
|
|
chr6_+_17231172
|
13.127
|
|
Cav2
|
caveolin 2
|
|
chr16_-_18426446
|
13.050
|
|
Comt
|
catechol-O-methyltransferase
|
|
chr6_+_54631644
|
13.017
|
NM_026629
|
2410066E13Rik
|
RIKEN cDNA 2410066E13 gene
|
|
chrX_+_162676870
|
13.012
|
NM_001177961
NM_001177962
NM_023122
|
Gpm6b
|
glycoprotein m6b
|
|
chr3_-_85550130
|
13.012
|
NM_172682
|
Fam160a1
|
family with sequence similarity 160, member A1
|
|
chr3_+_87834761
|
12.914
|
|
|
|
|
chr6_-_23082901
|
12.788
|
NM_013930
|
Aass
|
aminoadipate-semialdehyde synthase
|
|
chr9_-_77956353
|
12.616
|
NM_023605
|
Fbxo9
|
f-box protein 9
|
|
chr11_-_101026888
|
12.588
|
|
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr2_+_149656518
|
12.559
|
NM_001085521
|
Tmem90b
|
transmembrane protein 90B
|
|
chr14_+_8835385
|
12.512
|
NM_025341
|
Abhd6
|
abhydrolase domain containing 6
|
|
chr11_-_88712812
|
12.507
|
NM_009648
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr14_-_55734213
|
12.378
|
NM_001003829
|
Jph4
|
junctophilin 4
|
|
chr7_-_151621960
|
12.344
|
|
Cttn
|
cortactin
|
|
chrX_+_148613496
|
12.156
|
NM_001005475
|
Iqsec2
|
IQ motif and Sec7 domain 2
|
|
chr10_-_88720534
|
12.128
|
NM_178773
|
Ano4
|
anoctamin 4
|
|
chr9_-_42924626
|
12.067
|
NM_001034863
|
Tmem136
|
transmembrane protein 136
|
|
chr1_+_137665517
|
12.002
|
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr11_+_78136690
|
11.982
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr1_-_72920776
|
11.962
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr10_-_127058199
|
11.879
|
NM_008512
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chr17_-_87665228
|
11.874
|
NM_139295
NM_176808
|
Mcfd2
|
multiple coagulation factor deficiency 2
|
|
chr1_+_135142673
|
11.842
|
NM_001160268
NM_182930
|
Plekha6
|
pleckstrin homology domain containing, family A member 6
|
|
chr3_+_96405063
|
11.826
|
|
Lix1l
|
Lix1-like
|
|
chr8_+_97296110
|
11.726
|
|
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr3_+_90330770
|
11.488
|
NM_001163525
NM_001163526
NM_025393
|
S100a14
|
S100 calcium binding protein A14
|
|
chr12_+_36719492
|
11.452
|
NM_011783
|
Agr2
|
anterior gradient 2 (Xenopus laevis)
|
|
chr13_-_69138229
|
11.407
|
|
Adcy2
|
adenylate cyclase 2
|